Our Research
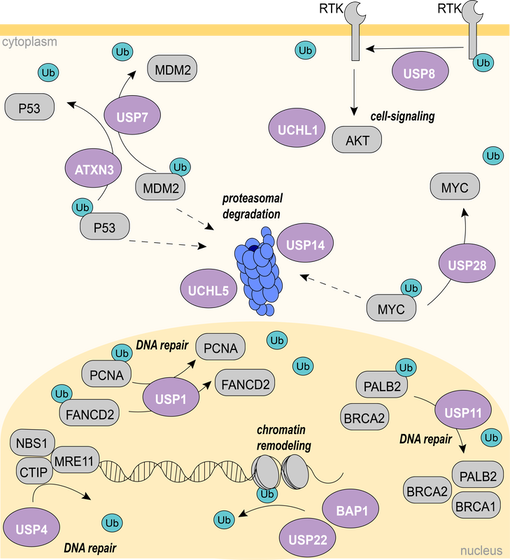 Buhrlage Lab - DUBs and their studied pathways
Buhrlage Lab - DUBs and their studied pathways
DUB inhibitor development
Deubiquitinating enzymes (DUBs) are peptidases that cleave the isopeptide bond between the C-terminus of ubiquitin and their substrate, removing ubiquitin from target proteins. Since the ubiquitin post-translational modification is essential for regulating protein degradation, activity, localization, and more, the ability to modulate DUB activity with small molecules has broad clinical and research applications. Therapeutically, DUBs have been implicated in diverse indications encompassing oncology, neurodegeneration, and infectious diseases. Excitingly, inhibiting DUBs offer the potential to degrade pathogenic proteins in a targeted manner.
DUBs are an emergent class of targets, akin to kinases 20 years ago. The first DUB-targeting agents are in the clinic and substantial progress has been made towards understanding DUB function. Moreover, DUBs are recognized as chemically tractable with multiple pockets for inhibitor, activator and recruiter development. Despite this, there are few high quality chemical probes for DUBs limiting ability to pharmacological investigate function and therapeutic potential.
We have built a DUB-focused chemical biology platform custom designed to accelerate development of selective probes for DUBs and annotation of DUB target biology. The integrated platform is comprised of DUB-targeted libraries with novel scaffolds and chemotypes and a suite of established and novel biochemical and chemoproteomic assays to profile compound activity and selectivity, as well as expertise in DUB medicinal chemistry, structural biology, computational chemistry, and functional annotation.
Deubiquitinating enzymes (DUBs) are peptidases that cleave the isopeptide bond between the C-terminus of ubiquitin and their substrate, removing ubiquitin from target proteins. Since the ubiquitin post-translational modification is essential for regulating protein degradation, activity, localization, and more, the ability to modulate DUB activity with small molecules has broad clinical and research applications. Therapeutically, DUBs have been implicated in diverse indications encompassing oncology, neurodegeneration, and infectious diseases. Excitingly, inhibiting DUBs offer the potential to degrade pathogenic proteins in a targeted manner.
DUBs are an emergent class of targets, akin to kinases 20 years ago. The first DUB-targeting agents are in the clinic and substantial progress has been made towards understanding DUB function. Moreover, DUBs are recognized as chemically tractable with multiple pockets for inhibitor, activator and recruiter development. Despite this, there are few high quality chemical probes for DUBs limiting ability to pharmacological investigate function and therapeutic potential.
We have built a DUB-focused chemical biology platform custom designed to accelerate development of selective probes for DUBs and annotation of DUB target biology. The integrated platform is comprised of DUB-targeted libraries with novel scaffolds and chemotypes and a suite of established and novel biochemical and chemoproteomic assays to profile compound activity and selectivity, as well as expertise in DUB medicinal chemistry, structural biology, computational chemistry, and functional annotation.
Visit our Publications page for recent published studies from the lab!
HOME | RESEARCH | PEOPLE | PUBLICATIONS | LAB NEWS | DIVERSITY | CONTACT
©2023 Cancer Chemical Biology at Dana-Farber Cancer Institute
©2023 Cancer Chemical Biology at Dana-Farber Cancer Institute
